Guest researcher blog post written by Sam Bryce-Smith – sharing his research into changes in ‘post-code’ sequences of RNA, how it continues during the COVID-19 pandemic, and the importance of his funding.
We are extremely grateful for the support of the London Freemasons, who are generously supporting one of our new PhD Studentships held by Sam at University College London (UCL), led by Dr Pietro Fratta and Dr Maria Secrier. The grant from London Freemasons comes through the Masonic Charitable Foundation, which is funded by Freemasons, their families and friends, from across England and Wales.
This PhD Studentship project investigates how RNA (molecules that carry information to make proteins from DNA) can be dysfunctional in MND by investigating the changes that occur using sequencing technologies.
About me
I am part of an ever-growing group of researchers who use and develop computer programs to improve our understanding of disease biology. My typical day-to-day involves using open-source, published programs to process and analyse RNA sequencing data generated by our lab, so I am very fortunate to be able to continue with my research remotely during the COVID-19 crisis (see header image of my work-from-home set-up!).
I have always been fascinated by how our bodies work. As my understanding of biology grew, I became ever more aware of how much we don’t understand about our biology and how it goes wrong. MND is a particularly striking example of this, so I wanted to contribute my efforts with the hope that this may help find therapeutic targets. Scientific research into the underlying causes of MND is sorely needed, and that is why I am honoured to have secured the Masonic Charitable Foundation Studentship. The importance of funding to understand the causes of MND should not be underestimated, as it provides the foundation for developing new treatments – by identifying the processes involved in MND, we can develop treatments that target these processes to stop MND in its tracks.
Background biology
DNA is the ‘cookbook’ of our cells – it contains a collection of distinct genes, or ‘recipes’, that contain the instructions to make distinct proteins, each of which has different jobs inside the cell. To make proteins, our cells first need to open our DNA and copy the gene into a messenger RNA molecule (mRNA).
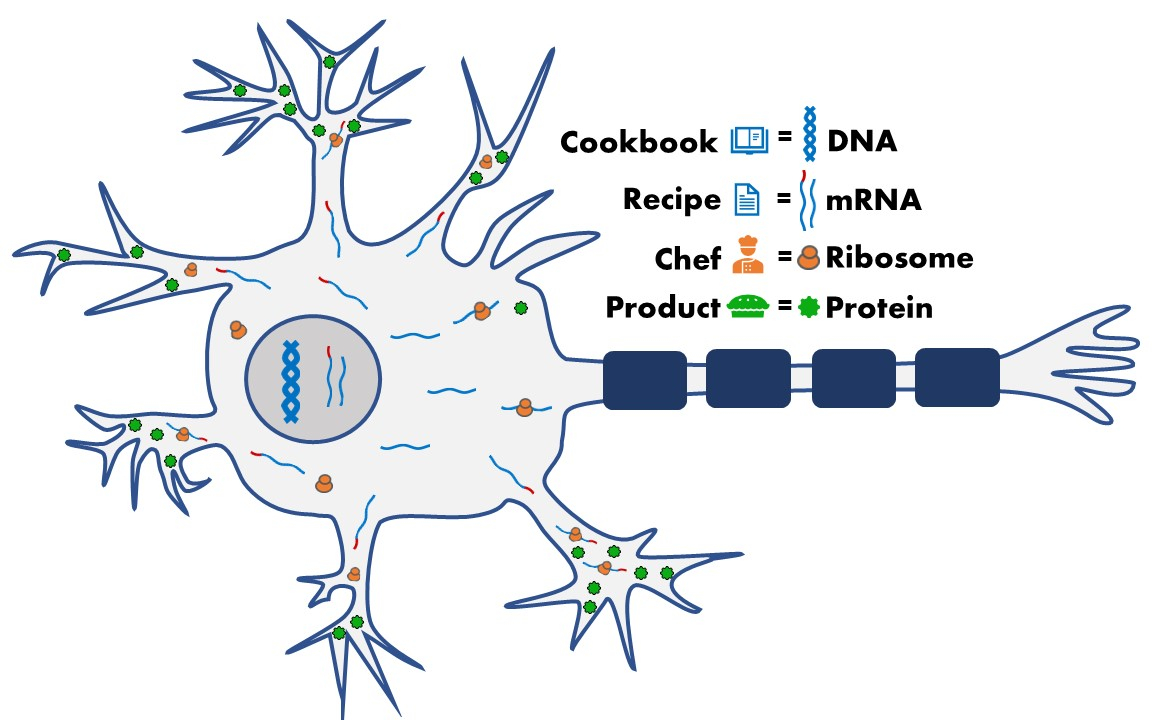
The mRNA doesn’t only contain instructions to make protein. Areas at the ends of mRNAs can contain ‘post-code’ sequences that deliver the mRNA to specific parts of the cell, so that proteins are made where they carry out their job.
Motor neurons are much larger than your typical cell – a single motor neuron can be over 1 metre long! Proteins and other important cellular substances, including mRNAs, are transported to and from the cell body of a motor neuron to their nerve endings, and neurons achieve this in part by using post-code sequences at the ends of mRNAs. Neurons are particularly reliant on processes that make sure proteins are where they need to be – see previous blog post about axonal transport.
A process called ‘alternative cleavage and polyadenylation’ can determine whether post-code sequences are included in the recipe to make proteins. My project will explore whether this process affects MND, and whether removing these post-code sequences could cause motor neurons to stop working. We will focus on studying the effects of TDP-43, a key protein that has been found to be affected in most cases of MND.
Studying changes in RNA using RNA sequencing
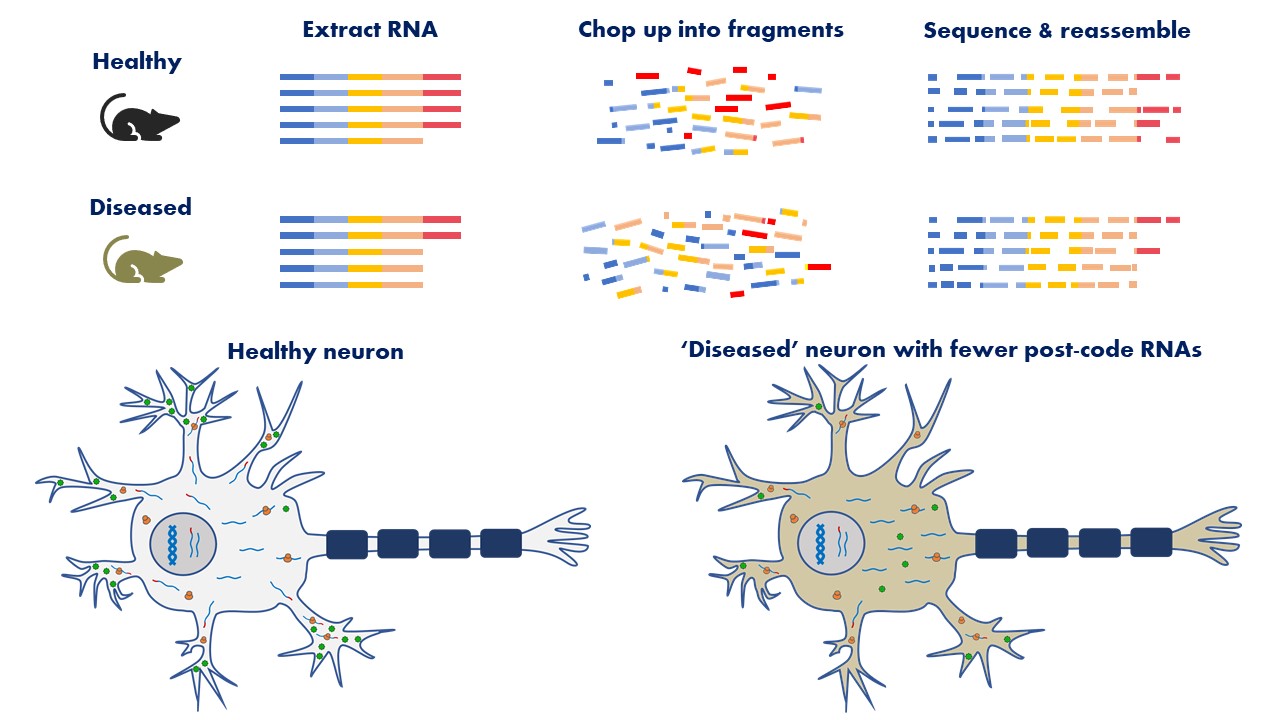
RNA consists of four different chemicals – adenine (A), cytosine (C), guanine (G) and uracil (U) – which form the ‘alphabet’ of the mRNA recipe. Different mRNAs have different combinations of letters, which are ‘read’ to produce different proteins. RNA sequencing determines the order of the letters in each RNA molecule, allowing us to study the differences between healthy and diseased neurons.
Current technologies cannot accurately sequence whole RNA molecules, so we chop up the RNAs into little pieces and sequence them individually. Like solving a jigsaw puzzle, we must then reassemble the pieces. RNA is a copy of a DNA sequence, so we use a standardised DNA sequence to help identify which gene the RNA comes from, like using the picture on the box to help assemble a jigsaw. A typical RNA sequencing experiment produces millions of sequence pieces, so we use computers programs to solve the puzzle. RNA sequencing has become a very important tool in our efforts to understand the underlying causes of MND.
Circling back to TDP-43
TDP-43 is a key protein in MND. It is usually found in the nucleus, where it is involved in producing mRNA (the copy of the DNA recipe). In MND, there are lower amounts of TDP-43 in the nucleus, which affects its ability to carry out all normal jobs. Previous research has suggested that TDP-43 may be involved in the ‘alternative cleavage and polyadenylation’ process which decides where the mRNA recipe finishes. If the recipe finishes too early, then post-code sequences at the end of RNAs could be removed and mRNA wouldn’t be delivered to the right place for the protein to be made.
To understand how this could affect a neuron, think of it like leaving the address off the front of an envelope. The letter still makes its way to the ‘depot’ (as the mRNA leaves the nucleus to the cell body), but with no address (post-code sequences), there is no way to know where the letter should be sent (‘postman proteins’ are not given the full mRNA recipe to deliver to the nerve endings). Without the full mRNA sequence to make the right proteins where needed; nerve endings may begin to stop working properly.
Summary
In my project, RNA sequencing will be to study how the levels of post-code sequences change between ‘diseased’ and ‘healthy’ neurons. I hope my research can consolidate our understanding of how TDP-43 contributes to MND. If we identify the processes that cause MND, treatments that target these processes can be developed that are more likely to be successful in stopping MND.
I feel very fortunate to be studying at the Institute of Neurology at University College London, a world-class centre for MND research. In just the first few months I feel my skills have come on leaps and bounds, and I am excited to continue my development and contribute to MND research in the long term.

We’d like to thank Sam for sharing his project through this blog post. We would also like to thank the London Freemasons for their generous support of this project.
- Read more about the research we fund in 2020 in our information sheet here.






Good man! A very informative and well-written post. As an MND sufferer i deeply appreciate the work you are doing, and wish you luck with your efforts.
Dear Hugh,
Thank you for taking the time to read and comment on our blog. Glad you enjoyed the post.
Research into MND continues to develop understanding of the disease. The early career researchers of today, such as Sam, may well be involved in finding more effective treatments, and maybe a cure, in the future.
Best wishes,
Research Development Team