ALS is the most common form of MND and these terms are used interchangeably in this blog.
MND is a complex disease, with many potential environmental and genetic factors that could lead to its development. Because of these underlying genetic causes, one promising option for treatment is gene therapy. Gene therapies target mistakes within the DNA of specific genes known to cause disease, and their use has been increasing in the past few years. Just recently, a gene therapy was approved to treat SOD1 MND in the United States, known as Tofersen. Advancements like these make it clear that more needs to be uncovered about the genetics behind MND to discover additional genes to target for gene therapy.
Unfortunately, the development of gene therapies is incredibly time-consuming and expensive. One way to fight against these burdens is to create multi-purpose drugs; medicine that can be administered for more than one condition, which could help more people living with neurodegenerative diseases get access to potential new treatments sooner.
In order to create multi-purpose drugs, diseases which share common genetic factors need to be identified. One recent paper has set out to do this with four neurodegenerative diseases. Using mathematical modelling, scientists were able to compare ALS, Parkinson’s disease, Lewy body dementia, and Alzheimer’s disease in an effort to discover any genetic links between them.
Overall, scientists were able to show ALS was most genetically linked with Alzheimer’s and identified three genes implicated in ALS that were also found in other neurodegenerative diseases. These results provide limited but interesting insights into potential targets for novel treatments that could benefit multiple conditions.
RELATED POST
Blog | 26 April 2023 | Charlotte Roy
What does the FDA approval of Tofersen mean for the UK MND community?
What did the study find?
The paper first looked at data from four large studies of each condition to determine whether any of the genetics behind the diseases were linked. They specifically looked at heritability, which is the proportion of genetic variation that can be attributed to a particular disease.
Genes, which are short sections of DNA, help to determine our individual characteristics. Genes also act as instructions to make proteins that carry out the many biological tasks our bodies require. Between individuals, the DNA that makes up genes can differ slightly. And while most of these differences are harmless, some can increase a person’s risk to diseases like ALS. These changes in DNA, that impact a person’s risk of developing a disease, are known as genetic risk variants. Heritability studies can measure how much of your DNA can affect your chances of developing complex traits, with neurological disorders being a prime example of a complex trait.
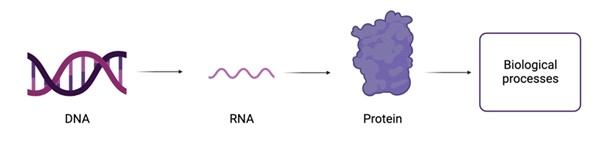
In order to create proteins, the building blocks of our body, a stretch of DNA first has to be copied to make an RNA molecule, which is then coded into proteins. These proteins are the building blocks of the body and are needed for a lot of biological processes within the body.
In the study, scientists found ALS had a weak but significant association with Alzheimer’s disease. This indicates some overlap of genetic risk variants between these two diseases. As it is already well documented that frontotemporal dementia shares genetic architecture with both ALS and Alzheimer’s disease, it is unsurprising that the two diseases are shown here to have an association.
The results also exhibited a weak association between ALS and Parkinson’s disease and a moderate association between ALS and Lewy body dementia. However, these results were not statistically significant so will require further analysis to confirm a correlation.
RELATED POST
Blog | 20 April 2023 | Eleanor Green
Measuring cognitive and behavioural changes in MND
What were the three genes of interest?
The paper then analysed the genetic risk variants associated with each disease by using mathematical modelling to see if there were any correlations between the genes involved in the neurodegenerative diseases. This identified three genes that overlapped with ALS and at least one other neurodegenerative disease.
One risk variant, in the gene TMEM175, was found to be significantly correlated with ALS, Parkinson’s disease, and Lewy body dementia. The TMEM175 gene is responsible for the formation of a protein that acts as a channel in and out of the lysosome, a sac-like organelle in the cell that breaks down proteins, carbohydrates, and lipids. Mutations in this gene have been shown to make the lysosome overly acidic, stopping the pathways responsible for the breakdown of proteins from functioning properly. This leads to a build-up of toxic protein deposits, also known as amyloids, that are thought to cause neurone degeneration.
Another variant, in the gene TNIP1, was found to be significantly correlated with both ALS and Alzheimer’s disease. TNIP1 is involved in pathways associated with the immune response and inflammation. The role of TNIP1 in both ALS and Alzheimer’s disease remains unclear, with scientists currently theorising that risk variants in this gene can lead to an accumulation of amyloid in the brain that becomes toxic. While there have been previous attempts to correlate specific regions in this gene to both diseases, they have been unsuccessful until this paper.
Finally, variants in the HLA gene regionwere found to be significantly correlated with ALS, Parkinson’s disease, and Alzheimer’s disease. HLA related genes are responsible for proteins that make up the human leukocyte antigen complex, an important component in the immune system that recognises foreign bodies like bacteria and viruses. Previous studies have confirmed that HLA related genes have significant risk variants in all three diseases. An unpublished study has even identified an HLA variant that is potentially protective in all three diseases. This indicates that HLA has a significant role in neurodegenerative diseases and could be a good candidate to target when developing potential new treatments.
Moving forward
While this research provides new insights into the genetics of neurodegenerative diseases, it did have some limitations. One major limitation was that the Lewy body dementia data had a very small sample size. Only around 2600 cases of Lewy body dementia were analysed compared to the 10,000+ cases of the other three diseases, making results from it less statistically powerful. If a larger sample size is analysed in the future, it is possible scientists would be able to find more significant overlap in both heritability and specific genetic variants between Lewy body dementia and ALS.
Moving forward, this research could open the door for development of gene therapies that could treat multiple neurological diseases. It could also provide insight into the underlying mechanisms behind these diseases and potentially be used to design genetic screens that cover multiple conditions. This could lead to increased scientific collaboration between multiple research groups, allowing for faster drug development and earlier diagnosis.